网站维护
系统内容更新/升级中

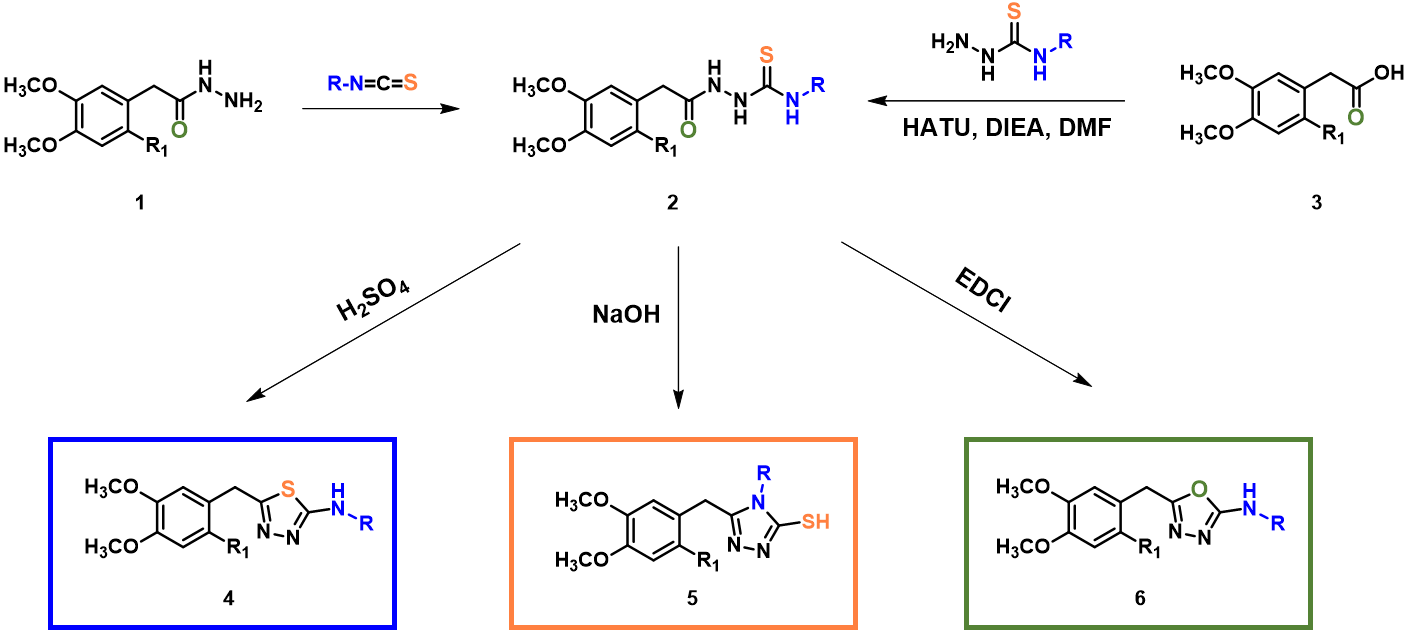
In organic synthesis, we encounter interesting substrates which under different reaction conditions provide different products. For example, reaction of the common intermediate 2, formed by reaction of acyl hydrazide 1 with isothiocyanate or by coupling of carboxylic acid 3 with hydrazinecarbothioamide (Figure 1), under acidic, basic, and neutral conditions, provided thiadiazole 4, triazole 5, and oxadiazole 6, respectively[1,2,3]. In this chapter, we’ll analyze these reactions with QM.
In organic chemistry, mechanistic insights are crucial in guiding our QM analyses: what are the structures/factors that could determine reactivities observed, what to calculate, how to calculate (Chapter 31).
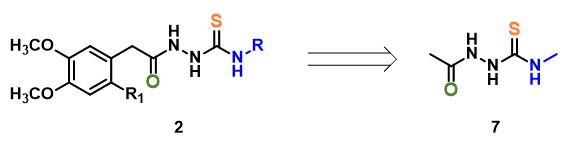
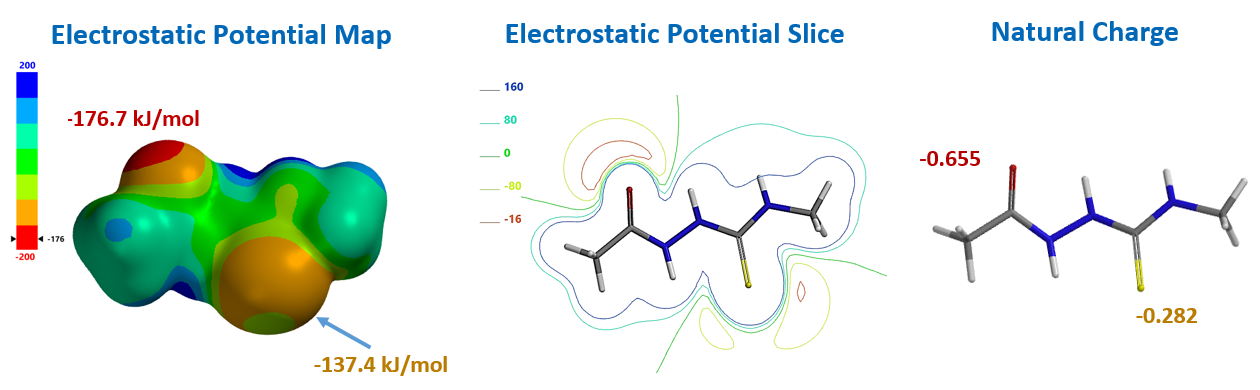
For computational efficiency, we simplified intermediate 2 as 2-acetyl-N-methylhydrazine-carbothiamide 7 (Figure 2). For acid mediated reactions, we need to find out which atom on 7 could be selectively protonated. We chose Electrostatic Potential Map and Natural Charge for the analysis[4]. With electrostatic potential map, by convention, colors toward red correspond to negative potential (stabilizing interaction between the molecule and a positive charge, eg. proton), while colors toward blue correspond to positive potential.
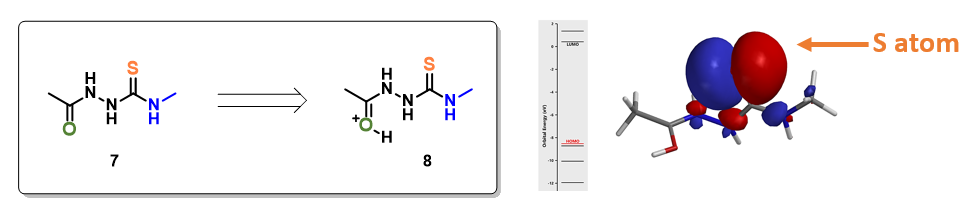
For structure 7, the most electronegative area (-176.7 kJ/mol) is on the carbonyl oxygen (Figure 3), consistent with the more negative -0.655 unit Natural Charge calculated for it. Electrostatic potential could also be displayed as two dimensional contour plot (slice). As such, preferentially protonation leads to intermediate 8. Calculation indicated that its HOMO mainly distributed on the sulfur atom, the nucleophilic center of the intermediate (Figure 3).
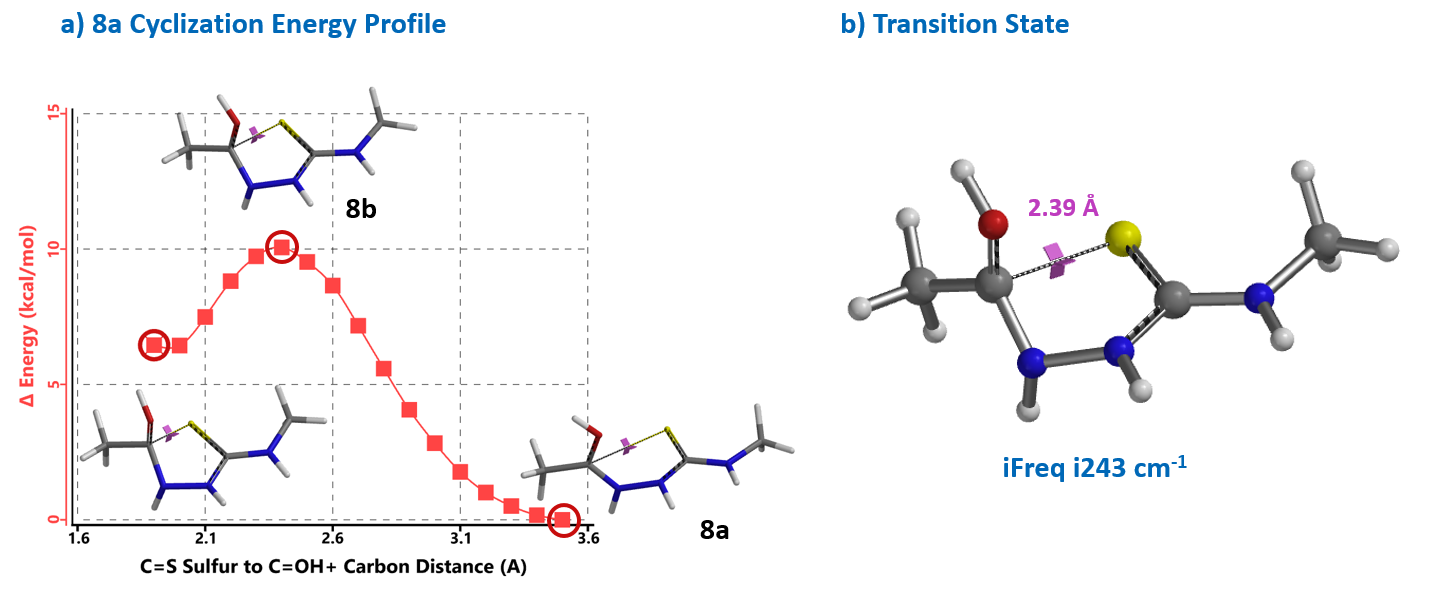
Reaction energy profile calculated for the cyclization step showed an activation energy of 10 kcal/mol (Figure 4), consistent with the observation that the reaction proceeds at 0 ºC. Further Transition State calculation with structure 8b showed only one imaginary frequency, i243 cm-1, which corresponds to the vibration of the C-S bond forming between the carbonyl carbon and sulfur atoms (Chapter 24).
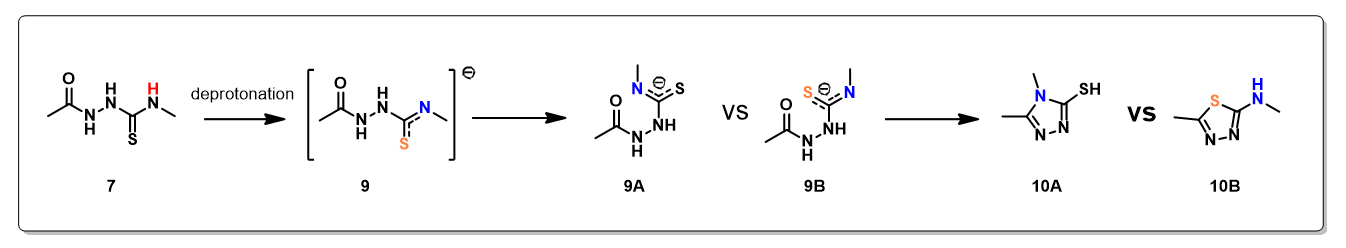
Electrostatic Potential Map revealed that the thioamide NH on structure 7 is the most acidic proton. Base deprotonation leads to anion 9, with the negative charge delocalized among the N-C-S atoms (Figure 5).
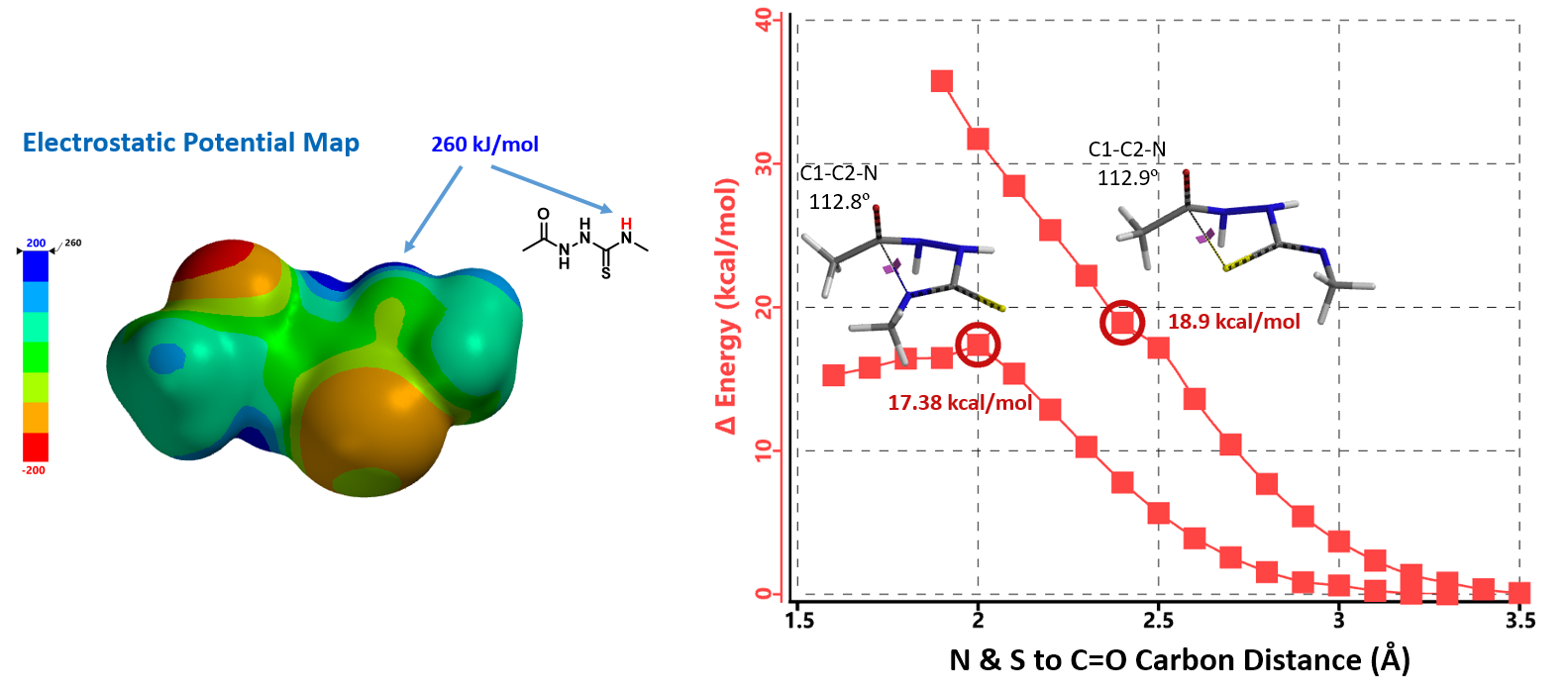
Reaction energy profile calculated for formation of triazole 10A suggested an activation energy of 17.38 kcal/mol, consistent with reaction temperature of 50-100 ºC required. For potential competing cyclization to thiadiazole 10B, calculated reaction profile is significantly higher in energy and continue to rise. This is consistent with our observations that only the triazole cyclization product 10A was formed.
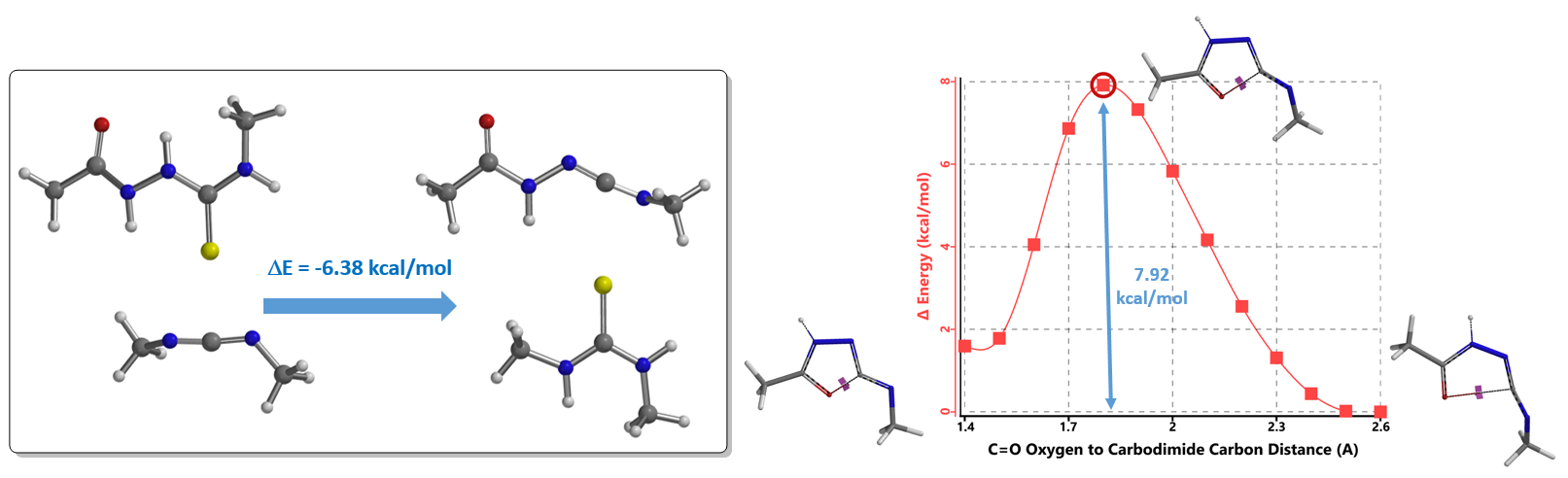
Several neutral conditions were reported for cyclization of structure 7 to oxadiazole. In this chapter, we’ll focus our discussion with the use of 1-(3-dimethylaminopropyl)-3-ethylcarbo-diimide (EDCI). We reasoned that the reaction is a two-steps process, hydrogen sulfide transfer from 7 to EDCI, and cyclization of resultant diimide 11 to oxadiazole 12. (Figure 6).
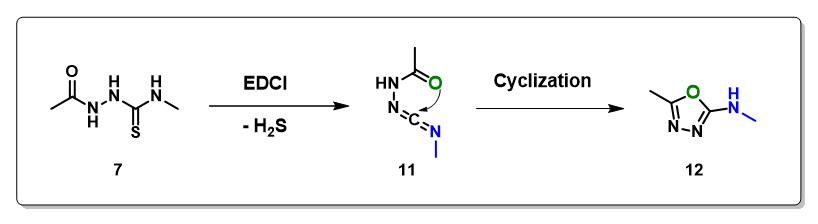
Calculations indicated that the hydrogen sulfide step is exothermic (-6.38 kcal/mol), and activation energy for the cyclization step is 7.92 kcal/mol. Both readily proceed.
QM analyses enable us to learn organic chemistry reactions at deeper levels. It requires us to be aware of the chemical properties of the substrates, reagents, reaction conditions, and reaction mechanisms, to find out what relevant QM parameters to calculate for to correlate with experimental results, to incorporate what we learned into our workflow, to turn QM into prospective tools. All these build on one another, require persistent efforts, to learn and understand the reactions of our interest, one at a time. Enjoy.
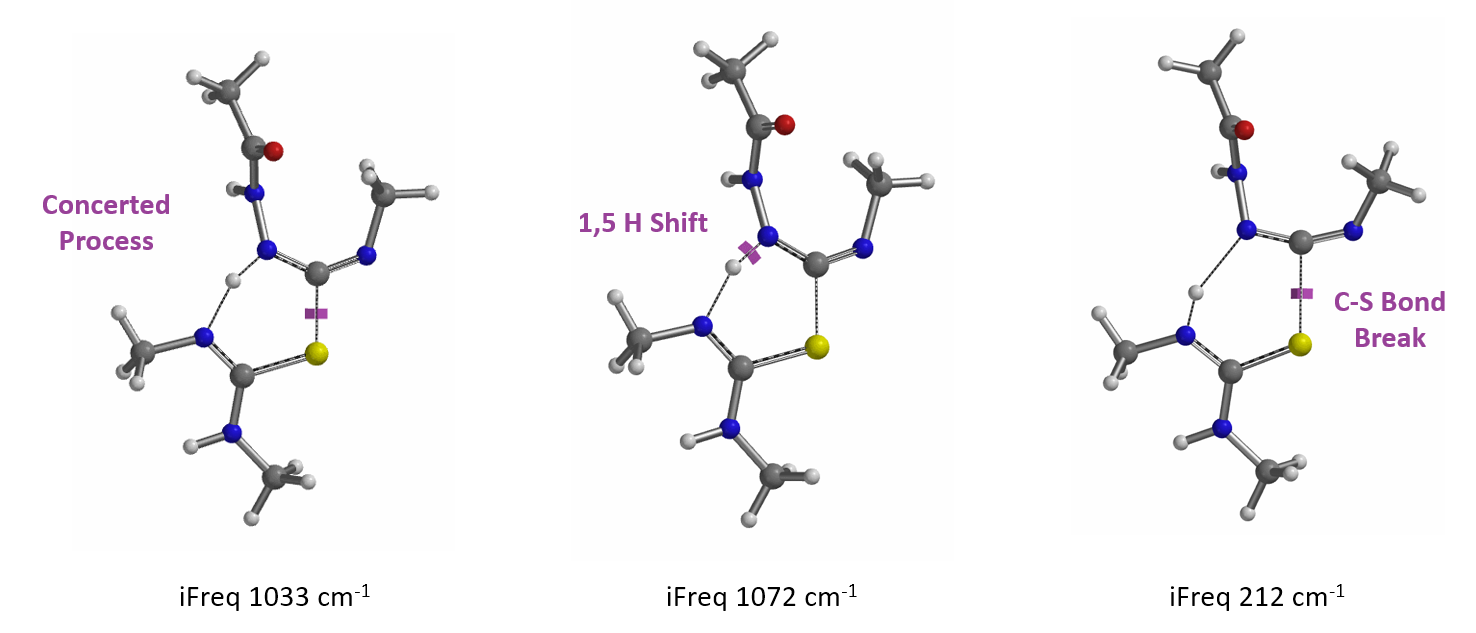
In the hydrogen sulfide transfer step discussed above, for the conversion of 13 to 11, (Figure 8), which mechanism is more consistent with QM calculation results? A concerted process or stepwise one?
This article is written and edited by Qiuyue Wang, Shouliang Wang, Yongsheng Chen, John S. Wai
References:
[1] P. Zoumpoulakis, C. Camoutsis, G. Pairas, M. Sokovic, J. Glamoclija, C. Potamitis, A. Pitas, Bioorg. Med. Chem. Lett, 2012, 20, 1569.
[2] I. Saramet, A. Banciu, L. Socea, C. Draghici, M.D. Banciu, Heterocyclic Communications, 2003, 9, 653.
[3] F. Naaz, F. Ahmad, B.A. Lone, Y.R. Pokharel, N.K. Fuloria, S. Fuloria, M. Ravichandran, L. Pattabhiraman, S. Shaf, M.S. Yar, Bioorganic Chemistry, 2020, 95, 103519.
[4] Spartan’20 Tutorial and User’s Guide (2020). Irvine, CA, USA: Wavefunction, Inc. p599; A.E. Reed, R.B. Weinstock, F. Weinhold, J. Chem. Phys. 1985, 83, 735; T. Lu T, F.W. Chen, Acta Phys.-Chim. Sin. 2012, 28, 1.