网站维护
系统内容更新/升级中

2020/05/07
DNA-encoded libraries (DEL) have become a significant tool for the hit identification in drug discovery. DEL technology allows the rapid construction of large chemical libraries of typically millions to billions of compounds, which can be quickly interrogated for target binding by affinity-based selection procedures followed by decoding with high-throughput DNA sequencing.
In most cases at the molecular level, the DEL diversity is confined to available building blocks of DNA-compatible chemical reactions. At this stage, a general reaction protocol with broad scope for the direct synthesis of DNA-conjugated aminomethylated arenes. Dr. Xuanjia Peng (WuXi DEL) and Prof. Xiaojie Lu (Shanghai Institute of Materia Medica) have reported a palladium-mediated Suzuki-Miyaura cross-coupling reaction of potassium aminomethyltrifluoroborate with DNA-conjugated aryl bromides on Biochem. Biophys. Res. Commun. recently. This novel DNA-compatible reaction proceeded well with excellent functional group tolerance under mild reaction conditions.
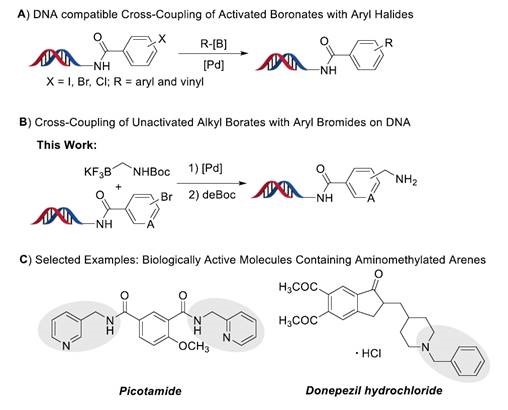
Suzuki-Miyaura cross-coupling reaction is a powerful tool for the construction of C-C bond and has become almost ubiquitous in drug discovery and medicinal chemistry. A number of Suzuki-Miyaura cross-coupling reactions have been developed towards DECL synthesis in the past years (Figure 1A). However, these DECL synthesis protocols have been limited: (1) Coupling partners are limited to boronic acids or esters (2) These DNA-compatible coupling reactions are limited to C(sp2)-C(sp2) bond formation. We reported the palladium-mediated coupling reaction of aminomethyltrifluoroborate with DNA-conjugated aryl bromides and C(sp2)-C(sp3) bond formation has been achieved for the first time (Figure 1B). Moreover, aminomethylated arenes occur in a variety of biologically active molecules, such as Picotamide and Donepezil (Figure 1C).
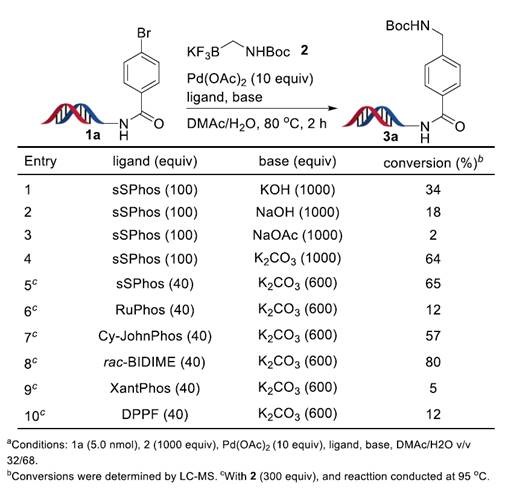
We started our investigation by examining the cross-coupling reaction between DNA-conjugated phenyl bromide 1a and potassium Boc-protected aminomethyltrifluoroborate 2 (Figure 2). When using Pd(OAc)2 (10 equiv) and sSPhos (40 equiv), K2CO3 (600 equiv.) was found to be the best inorganic base. The reaction gave the desired product 3a with 65% yield. The use of ligand RuPhos and Cy-JohnPhos afforded 3a in 12% and 57% yield respectively. (rac)-BIDIME was found to be most effective, giving 3a 80% yield.
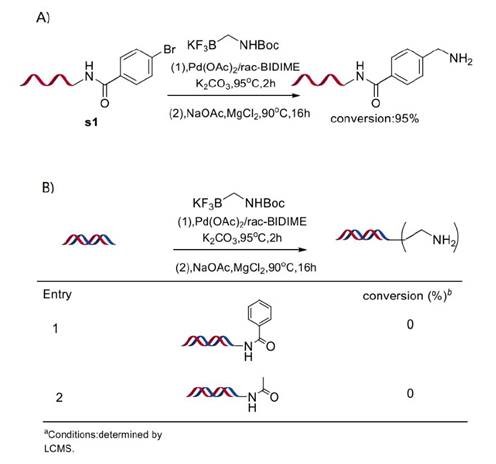
Although more susceptible to damage, ssDNA tags have been used for DEL construction. Excellent yield was obtained when using ssDNA-conjugated substrate s1 under the corresponding optimised conditions for dsDNA-conjugated substrates (Figure 3A). To confirm that coupling wasn’t occurring unexpectantly with the DNA oligomer itself, reactions between DNA-conjugated benzamide and acetamide were performed and no desired products were obtained (Figure 3B).
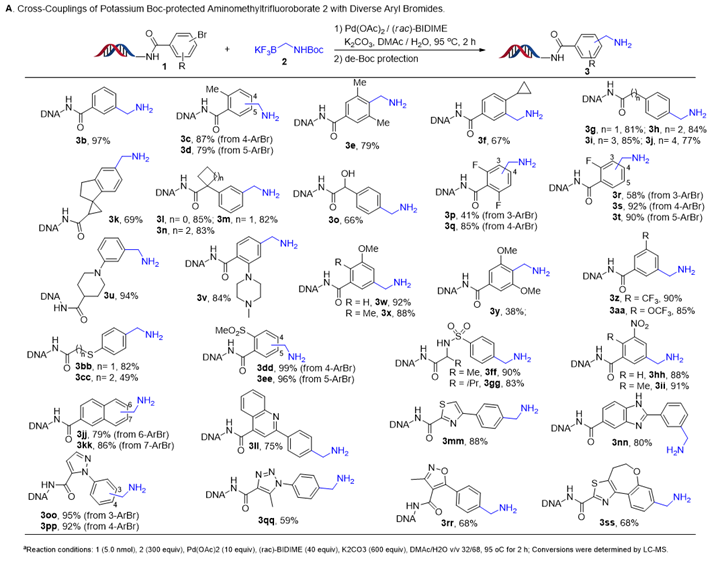
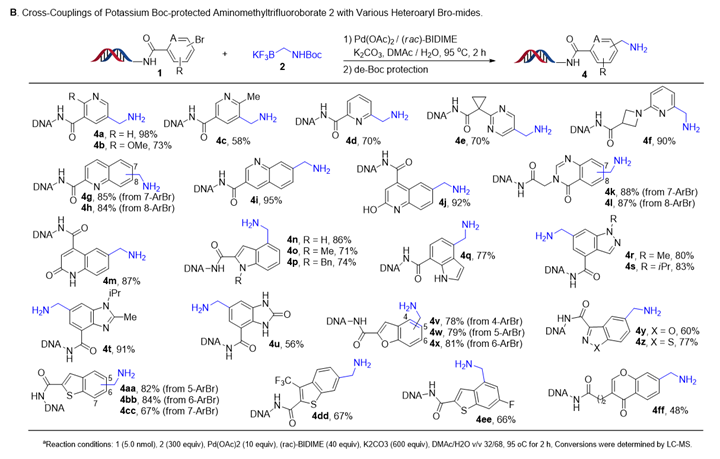
The substrate scope of the reaction was investigated under the optimised reaction conditions, followed by Boc deprotection. Initially, we focused on DNA-conjugated aryl bromide derivatives (Figure 4). Aryl bromides with either electron-withdrawing or electron-donating groups on the benzene ring were compatible. Functional groups, such as halides, ether, hydroxyl, thioester, sulphone, sulfamide, nitriles and amides, were all tolerated under the optimised reaction conditions. Extension of these conditions to aryl bromides with heterocyclic substituents, such as quinoline and oxazole also led to good conversions to product. Then we focused on the scope of DNA-conjugated heteroaromatic bromides (Figure 5). We also found that a variety of substrates containing indazole, benzimidazole, benzimidazolone, benzofuran and benzothiophene were compatible with the reaction conditions.
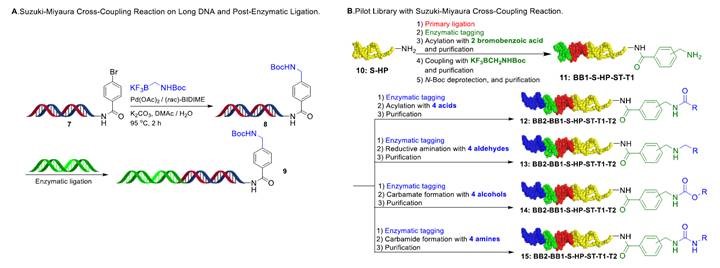
The cross-coupling reaction of modified elongated dsDNA substrate 7 with alkyltrifluoroborates 2 also afforded the desired product 8 in good yield. The enzymatic ligation of 8 with another dsDNA tag proceeded smoothly and the corresponding longer DNA product 9 were identified by the gel electrophoresis image (Figure 6A). Damage to DNA was quantified by NGS and qPCR experiments, indicating that no measurable DNA damage occurs upon the Suzuki-Miyaura cross-coupling reaction. DNA-conjugated aminomethylated arene products were able to undergo a variety of organic transformations, such as acylation and reductive amination, demonstrating its potential in DECL synthesis as a versatile intermediate (Figure 6B).
Summary
Dr. Xuanjia Peng (WuXi DEL) and Prof. Xiaojie Lu (Shanghai Institute of Materia Medica) have reported a palladium-mediated Suzuki-Miyaura cross-coupling reaction of potassium aminomethyltrifluoroborate with DNA-conjugated aryl bromides for the efficient synthesis of DNA-conjugated aminomethylated arenes. The utility of the methodology was demonstrated by conducting various transformations of DNA tagged aminomethylated arenes as would be done for a two-cycle, “split-and-pool” type DECL synthesis.
This work has been published on Biochem. Biophys. Res. Commun.
Palladium-Mediated Suzuki-Miyaura Cross-Coupling Reaction of Potassium Boc-protected Aminomethyltrifluoroborate with DNA-Conjugated Aryl Bromides for DNA-Encoded Chemical Library Synthesis
Reference:
Qu, Y.; Liu, S.; Wen, H.; Xu, Y.; An, Y.; Li, K.; Ni, M; Shen ,Y.; Shi, X.; Su, W.; Cui, W.; Kuai, L.; Satz, A. L.; Yang, H.; Lu, X;. Peng, X. Biochem. Biophys. Res. Commun. 2020. DOI: 10.1016/j.bbrc.2020.04.027