网站维护
系统内容更新/升级中

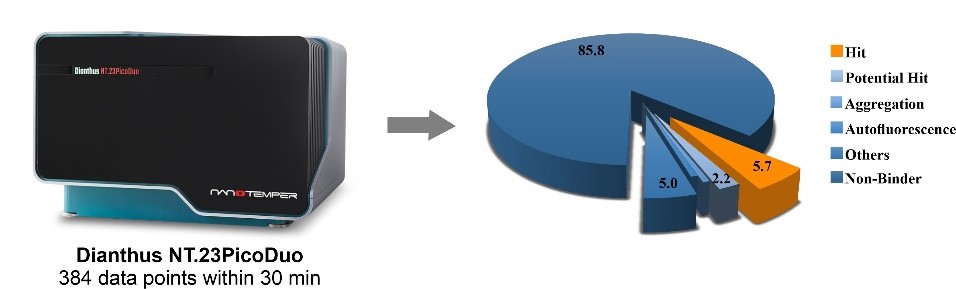
In the past, thousands of proteins were considered undruggable. The restricted number of chemical entities available and classical methods limited the exploration of this undevelopable area. Today, the combination of novel chemical modalities and advanced technical approaches has resulted in new clinical candidates from previously undruggable targets. To accelerate your drug discovery campaign, WuXi AppTec´s HitS subunit Crelux in Munich, Germany, has acquired the state-of-the art device Dianthus (NanoTemper Technologies GmbH), which enables the quantification of molecular interaction by measuring Temperature Related Intensity Change (TRIC). The measurements take place free in solution and in a microplate format. Usually the target is conjugated to the TRIC-optimized red dye. Because only a very brief laser-induced temperature change is applied, we can measure 384 data points within 30 min and up to 19 000 per day (Figure 1). This enables us to run and analyze the carefully tendered HitS Fragment Library of 3100 fragments or your own library at an unprecedented speed. The hits can then be confirmed using various biophysical methods including MST, SPR, ITC, nanoDSF and the protein X-ray crystallography.
Typical fragment screening workflow is depicted below based on the in-house STING screening campaign (Figure 2). Stimulator of Interferon Genes (STING) plays a crucial role in the innate immune system and was long perceived as a difficult and almost undruggable target.
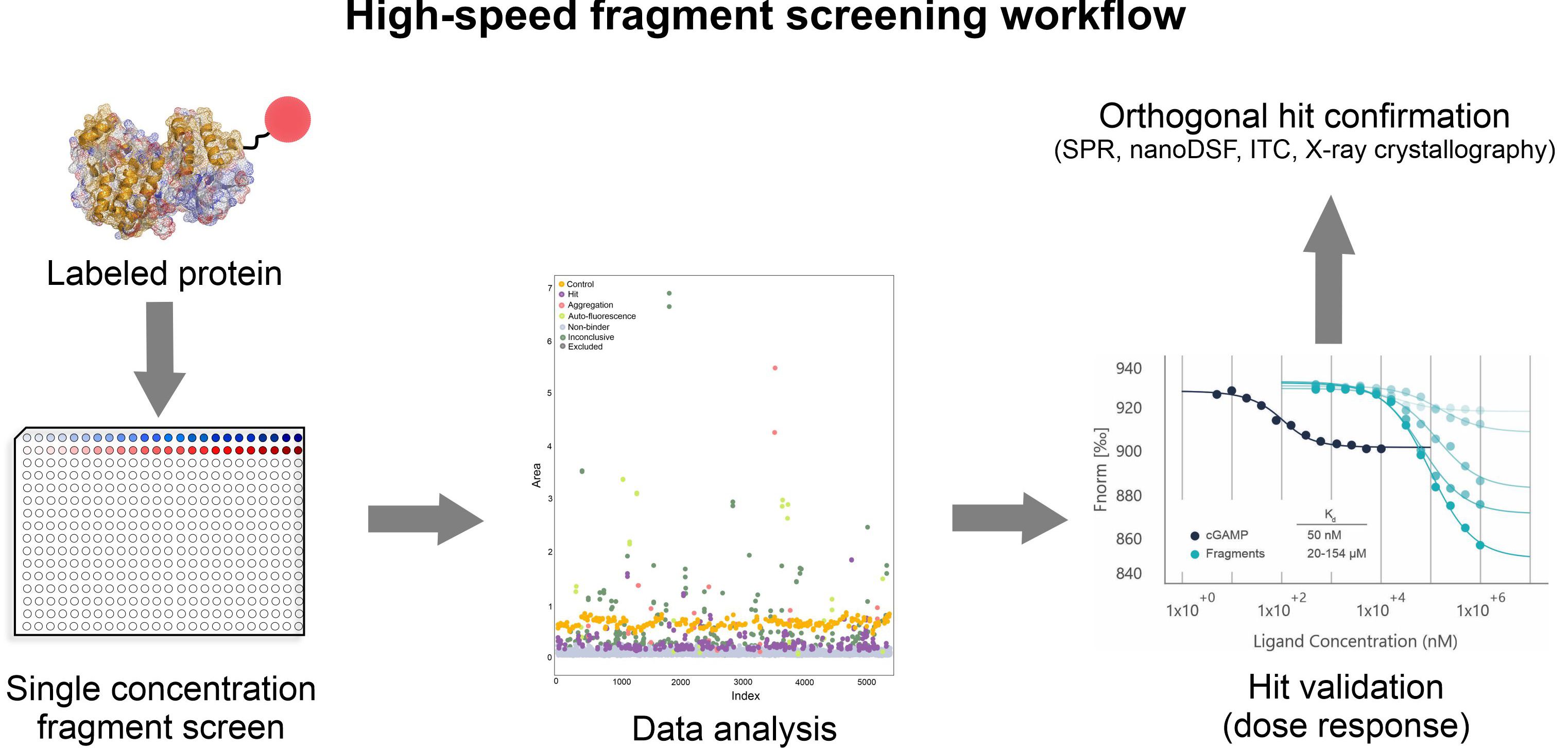
Our Protein Production department produced the crystal grade quality STING protein (the cyclic dinucleotide-binding domain (CBD)). Prior the screening, strict assessment of optimal screening conditions was performed. Various parameters like labeling conditions, protein stability (using nanoDSF), assay window and signal-to-noise ratio were monitored under different buffer conditions and over time. Dicyclic GMP was used as a positive control. After the stringent validation of assay conditions, interaction of 2213 fragments to STING was tested at the single concentration (0.5 mM, duplicate, measurement time 8 hours). 125 fragments were identified as hit, further 37 as potential hit. For these 162 fragments a full dose-response was measured and the Kd values determined. For 80 fragments, the affinity between 20 to 500 µM was determined. Top 66 fragments were additionally characterized and their binding to STING confirmed with orthogonal methods SPR and nanoDSF. As the last step of the hit confirmation process we intend to obtain the structural information by using X-ray crystallography and determine the fragment binding sites and the orientation of fragments in the binding pocket (Figure 3, STING bound to di-cGMP).
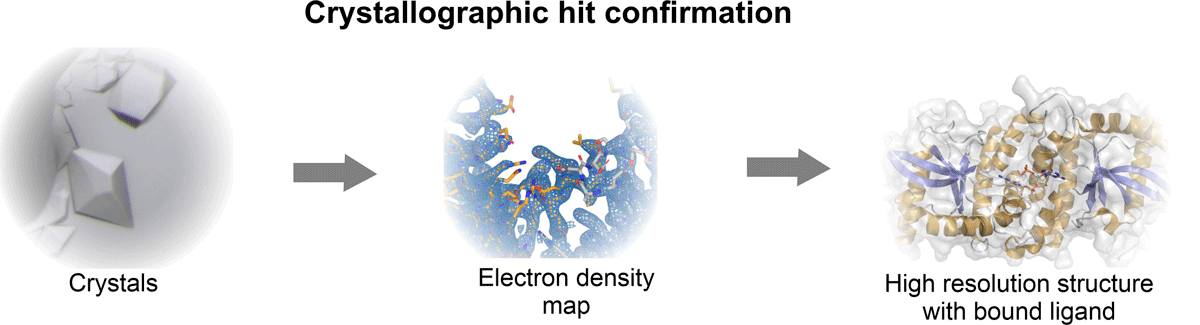
The X-ray crystallography is the most widely used method for structural hit confirmation that provides a high resolution of the fragment-binding crevice. Throughput with established targets cannot be matched with any other method; the resolution and level of detail that can be achieved is currently unsurpassed. These assets of X-ray crystallography enable fast processing of the fragment hits to produce the co-structures. This provides valuable data to support and accelerate in silico fragment-to-lead development and medicinal chemistry design.
Carefully designed protein construct, strict assessment of screening assay conditions, unprecedented speed of screening enabled by Dianthus combined by the robust hit validation and structural hit confirmation are the strengths of the fastest fragment screening accessible on the market and provided by WuXi AppTec´s HitS.
Contact us for more information at: HitS_service@wuxiapptec.com.