网站维护
系统内容更新/升级中

2020/05/19
DNA-encoded library (DEL) is successful for hit identification and widely used in drug discovery. Machine learning is particularly valuable and promising towards DNA-encoded library (DEL) technology as its ability in building blocking selection.
Recently, Dr. Xuanjia Peng (WuXi AppTec DEL), Prof. Xiaojie Lu (Shanghai Institute of Materia Medica) and Prof. Mingyue Zheng (Shanghai Institute of Materia Medica) have reported a novel DNA-compatible Pictet-Spengler reaction aided by a machine learning algorithm on iScience.1 This DNA-compatible reaction proceeded well with good functional group tolerance under mild reaction conditions. Building blocks selected by our trained machine learning algorithm have a higher pass rate (79.4% building blocks yielded more than 50%) than selected in random (18.4% building blocks yielded more than 50%). This is the first demonstration of using a machine learning algorithm to cull potential building blocks prior to their purchase and testing for DNA encoded library synthesis.
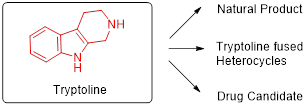
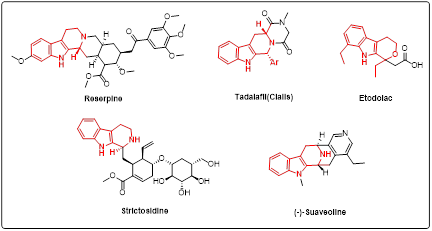
Initially, we focused on the DNA compatible cyclisation of highly functionalised and rigid rings. Poly-substituted optically active tryptoline derivatives are common structural motifs in indole-based alkaloids and usually prepared by Pictet-Spengler reaction. Functionalisation of the C-1 position of tryptoline derivatives is generally observed in natural-product-based indole alkaloids and commercial drugs (Figure 1). Under optimised reaction conditions, a majority of randomly chosen aldehyde building blocks fail to give high conversation to desired products. To better filter commercial building blocks, we trained a deep neural network (DNN) model to predict the conversion rate of building blocks in the reaction. We then purchased a subset of these building blocks and compared our model’s predictions with experimental results.
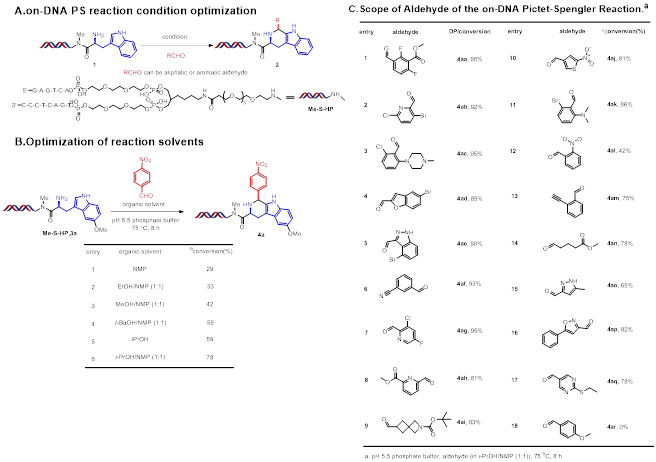
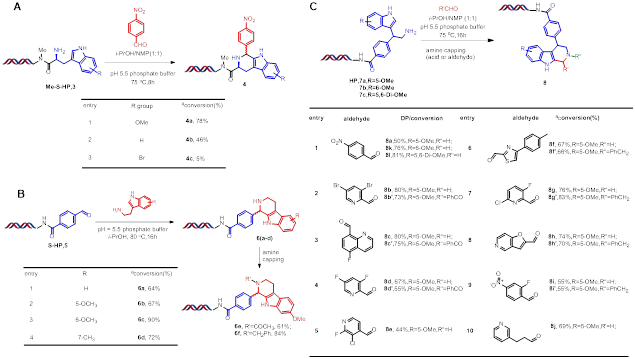
Reaction optimisation of this DNA-compatible Pictet-Spengler reaction was performed at the first stage using DNA-conjugated tryptamine substrates 1 (Figure 2A). We found that neither Lewis acids Sc(OTf)3, In(OTf)3, YbCl3, YCl3, Sm(OTf)3 nor Brønsted acids H3PO4, HCOOH appeared to promote the reaction and only DNA damage was observed. Basic conditions also did not promote the reaction, and using I2 also gave disappointing results. Then we chose to investigate a methoxy substituted tryptamine conjugated DNA substrate 3a. Employing a pH 5.5 phosphate buffer to maintain a weakly acidic condition, we observed 29% conversion to 4a and no obvious signs of DNA damage. We also screened a series of reaction solvents (Figure 2B). The solvent iPrOH led to an increase to 59% conversion. Using a 1:1 mixture of NMP and iPrOH gave an increase in conversion to 78%. With the optimised reaction conditions in hand, we explored the substrate scope of this DNA-compatible Pictet Spengler reaction (Figure 2C). The reaction proceeded well with a number of aldehydes. Functional groups such as halides, esters, alkynes and nitriles were tolerated. Heterocyclic aryl aldehydes gave moderate to excellent conversion.
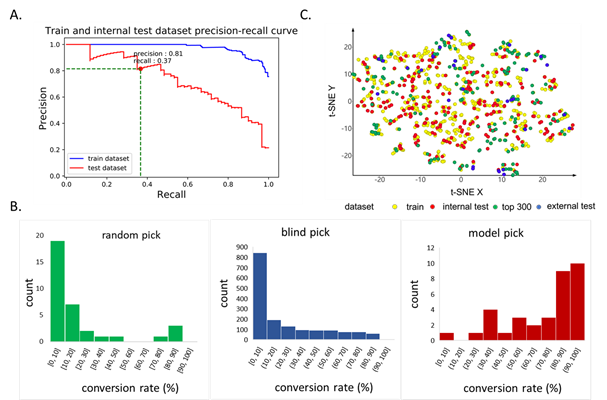
A useful model must correctly identify building blocks that latter give high conversion rate in the test PS reaction. To better quantify the performance of the model, building blocks with conversion rates over 50% are labeled 1, and others are labeled 0. Following this definition, a precision-recall curve can be plotted (Figure 3A). The results indicated a satisfactory performance for selecting building blocks with high conversion rates. The performance of the model on the external dataset was similar to that on the internal test dataset, with a precision for identifying building blocks with conversion rate above 50% being 0.79 (0.81 for the internal test dataset). Compared with blindly picked building blocks, the model has a better performance to find high conversion rate building blocks (high conversion rate BBs percentage: 18.4% vs 79.4%, Figure 3B). In practice, more rigorous clustering selection can be made to make picked building blocks as diverse as possible if enough building blocks are available. Our model could be served as a proof-of-concept in how building blocks can be filtered for purchase.
We investigated differing DNA-conjugated tryptamines. The electronic effect of the substrate had an important influence on the reaction, and unsurprisingly that the methoxy substituted substrate (3a) gave the best result while a bromosubstituted substrate gave almost no desired product (Figure 4A). Reactions between a DNA-conjugated aryl aldehyde and different tryptamine substrates proceeded smoothly with good to excellent conversions (Figure 4B). on-DNA PS reactions between DNA conjugated indole substituted amine 7 and different aldehydes were performed, with high yields and excellent functional group tolerance obtained (Figure 4C).
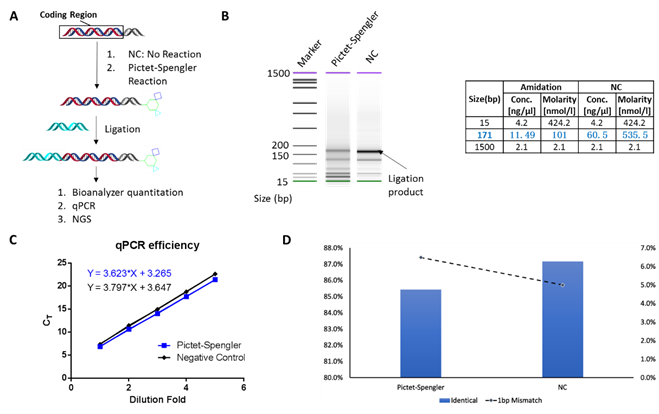
The comprehensive DNA integrity assessment study, such as NGS and qPCR, demonstrated no damage to DNA by the Pictet-Spengler reaction, and thus, could potentially be used for the DNA encoded library construction (Figure 5). We also performed proof-of-concept synthesis for two different three-cycle libraries, and these two diverse library designs demonstrate the potential of this novel on-DNA PS reaction (Figure 6).
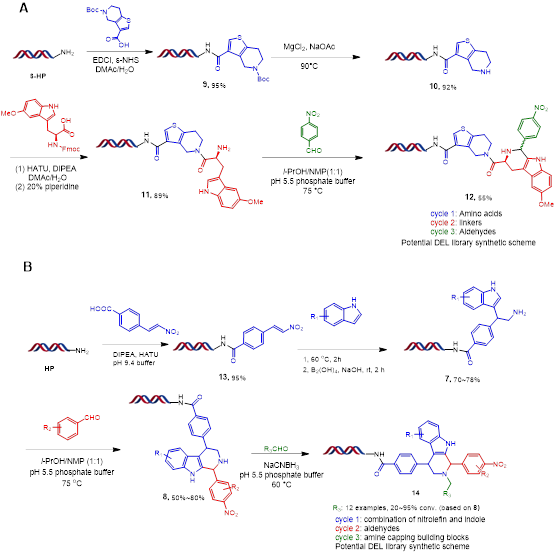
In summary, with the rational design of the on-DNA indole substrates, we have developed the first DNA-compatible Pictet-Spengler reaction for a variety of aldehydes. Suitable reaction conditions were identified for various combinations of PS reaction coupling partners. A DNN model has been developed to make the prediction of the reaction conversion rate for the building blocks, which was the first example of applying machine learning for the building block selections of the corresponding on-DNA reactions for DNA encoded library synthesis.
References: